Elena Pourmal and Quincey Koziol – The HDF Group
UPDATE: Check our support pages for the newest version of HDF5-1.10.0.
This version contains a fix for an issue which occurred when building HDF5 within the source code directory. Check our downloads page for more information. We are still on target for releasing HDF5-1.10.0 next week, let us know if you have any comments! |
The HDF Group is committed to meeting our users’ needs and expectations for managing data in today’s fast evolving computational environment. We are pleased to report that the upcoming major new release of HDF5 (HDF5 1.10.0) will have new capabilities that address important data challenges faced by our community. In this blog we introduce you to some of these exciting new features and capabilities.
More powerful than ever before and packed with new features, the release is scheduled for March, 2016. Among many enhancements, HDF5 1.10.0 addresses:
| Area of enhancement | New feature |
| HDF5 performance when appending data | Scalable chunk indexing |
| HDF5 compression | Enabled contiguous dataset compression |
| Use of free space in DHF5 files | Persistent free file space tracking |
| Access data while writing to HDF5 | Concurrent Single Writer/Multiple Reader |
| Access data across files and datasets | Virtual datasets |
| Scalability of parallel HDF5 | Multiple enhancements to parallel HDF5 |
If you have encountered challenges in any of these areas, then we are certain that the upcoming HDF5 1.10.0 will be of interest to you. We will be releasing these new features in the HDF5 1.10.0-alpha versions, starting in Fall 2015. We intend to blog about new features in more detail, but for now, here is an overview of what is coming in HDF5 1.10.0.
I/O Performance boost using scalable chunk indexing
One of the most powerful features in HDF5 is the ability to add more data to an existing data array stored in an HDF5 dataset. Those applications that just append data will especially benefit from this feature. As many of you may know, this feature requires a special data storage mechanism called chunk storage (see https://support.hdfgroup.org/HDF5/doc/Advanced/Chunking/index.html). However, the feature comes with some drawbacks that can affect performance. (We will discuss the steps users can take to boost performance in an upcoming blog.) There are several library enhancements to chunking that are coming in HDF5 1.10.0 which will help many applications get better performance results than with the HDF5 library version 1.8.
To look up chunks for each I/O request, chunked HDF5 datasets must have an indexing structure. Although HDF5 currently uses a B-tree for indexing chunks, which has a logarithmic time complexity, i.e. O(log n), HDF5 files and operations have a strongly structured nature. For the majority of the HDF5 community’s applications, this feature can be taken advantage of to create constant time chunk look ups, i.e. O(1).
We’re improving the indexing structure, which will offer many efficiencies and advantages:
- Zero storage cost for index when chunk locations are pre-defined;
- Less storage overhead for index and constant look-up time for a dataset with the fixed-size dimensions;
- Less storage overhead for index, and constant look-up and append time for a dataset with one unlimited dimension;
- Less storage overhead for index and performance for a dataset with multiple unlimited dimensions.
New data structures allow all forms of chunked datasets to be accessed in constant time, except those with multiple unlimited dimensions, an unprecedented performance boost for HDF5 applications.
Enabled compression for a contiguous dataset in this release
HDF5 compression is another powerful feature. In the HDF5 1.10.0 release we enabled compression (and other filters like checksum) for a contiguous dataset.
This feature improves performance of the applications that need to store compressed data and do not need access to the parts of the data. In this case the library writes and reads one contiguous block of compressed data, bypassing the lookups that would be needed for chunked storage.
Reuse free file space with persistent free file space tracking
From the initial release of the HDF5 library, HDF5 files have lacked persistent free space tracking. Although free space was tracked within a file while it remained open, there was no ability to allow space from deleted objects or resized compressed chunks to be reused when the file was closed. All that information (and space!) was lost to future applications that modified the HDF5 file. This led to stopgap measures such as the h5repack tool, for copying an HDF5 file and eliminating the unused space in the process.
With the HDF5 1.10.0 release, developers now can enable persistent free file space tracking for HDF5 files, allowing space within an HDF5 file to be reused by later writers.
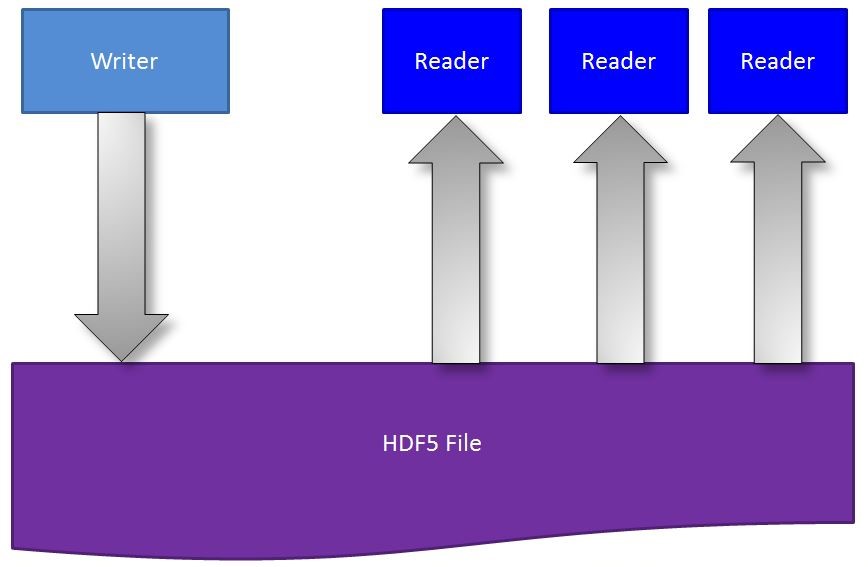
Concurrent read/write access with Single Writer/Multiple Reader (SWMR)
Data acquisition systems often need to analyze and visualize data concurrently while writing it, to determine if they need to take action immediately. Bringing such a capability to HDF5 would enable users with high bandwidth data streams to simultaneously store and monitor their data.
To support users in such environments, we have developed a capability for concurrent read/write access to HDF5 files that we call single-writer/multiple-reader (SWMR – pronounced “swimmer”) access. SWMR functionality allows a writing process to append data to datasets in an HDF5 file while several reading processes can concurrently read the new data from the file (Figure 1). No communications between the processes and no file locking are required. The processes can run on the same or on different platforms, as long as they share a common file system that is POSIX compliant. Click for more information on SWMR.
Virtual Datasets to access data across files and datasets
With a growing amount of data being produced in distributed architectures, a new need has emerged to access data stored across HDF5 files using standard HDF5 objects such as groups and datasets – without rewriting or rearranging the data.
An HDF5 virtual dataset (VDS) is an HDF5 dataset that is composed of source HDF5 datasets in a predefined mapping. Figure 2, below, shows a simple mapping of source datasets to a VDS, but much more complex mappings are possible. HDF5’s new virtual dataset capabilities will be particularly useful in an environment where multiple data sources need to update an HDF5 dataset simultaneously.
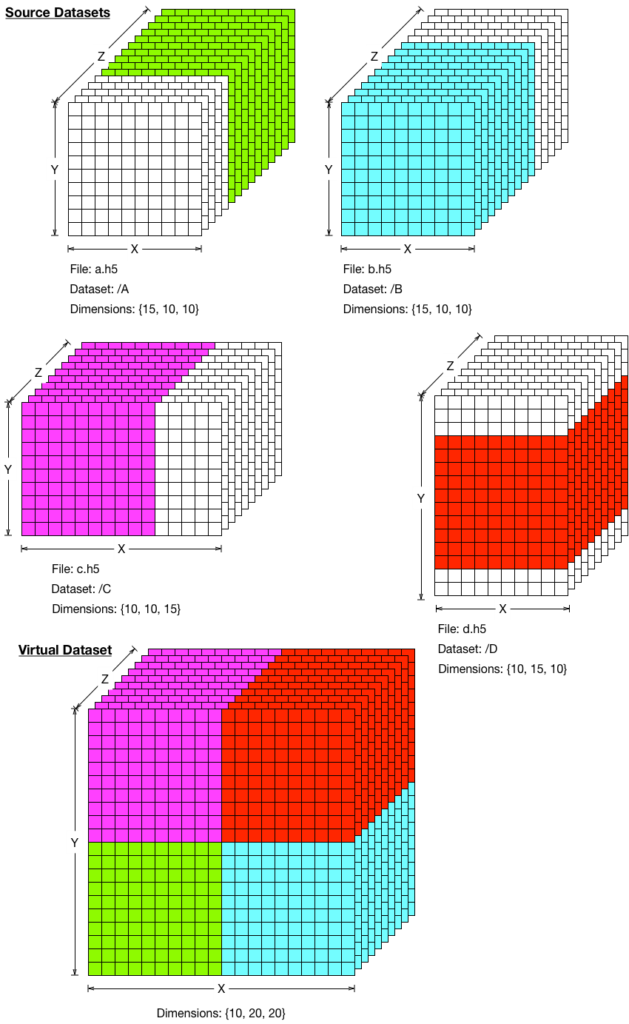
Virtual datasets can be useful, for example, at Synchrotron centers such as the UK’s Diamond Light Source (DLS) and Deutsches Elektronen-Synchrotron (DESY). These enormous particle accelerator facilities will be generating and storing terabytes of experimental data per day in HDF5 files. Because of the nature of the experiments and hardware constraints, the data representing, for example, an X-ray image, will be stored across different HDF5 datasets in multiple HDF5 files. The whole image can be accessed by an application without any specific knowledge of where data for each part of the image is stored.
The fact that a dataset is virtual is completely transparent to HDF5 applications. Users can access and work with data stored in a collection of HDF5 files in the same manner as if it were in one file. There is no need to change the way the data is collected and stored.
Enhancements to Parallel HDF5
Over time and as we work with more parallel HDF5 application developers, we have encountered and addressed several performance bottlenecks in parallel HDF5, leading to the new features below:
• Speed up file closing by avoiding file truncation: The HDF5 file format through the 1.8 release series has required that an HDF5 file be explicitly truncated when it is closed. However, truncating a file on a parallel file system can be an exceptionally slow operation. In response, we have tweaked the HDF5 file format to allow the file’s valid length to be directly encoded when the file is closed, avoiding the painful truncation operation.
• Faster I/O with collective metadata read operations: Metadata read operations in parallel HDF5 are independent I/O operations, occurring on each process of the MPI application using HDF5. However, when all processes are performing the same operation, this can cause a “read storm” on the parallel file system, with each process generating I/O operations that are identical across the application. We have addressed this problem by adding a collective I/O property to metadata read operations such as opening objects, iterating over links, etc. This allows a single process to read metadata from the HDF5 file and broadcast the information to the other processes, drastically reducing the amount of I/O operations the file system encounters.
• Improved performance through multi-dataset I/O: The HDF5 library now will allow a dataset access operation to access multiple datasets with a single I/O call. The new API routines, H5Dread_multi() and H5Dwrite_multi(), perform a single I/O operation to multiple datasets in the file. These new routines can improve performance, especially in cases when data accessed across several datasets from all processes can be aggregated in the MPI-I/O layer. The new functions can be used for both independent and collective I/O access, although they are primarily focused on collective I/O.
Taken together, these improvements to parallel HDF5 can significantly improve the performance of most, if not all, parallel HDF5 applications, generally with minimal changes to the application.
In addition to the major features described above, virtually every aspect of the HDF5 library has been updated and optimized so it can more easily be maintained and extended in the future.
Faster, more powerful, scalable, flexible and efficient, HDF5 1.10.0 is a major new release with unprecedented performance boosts and advanced capabilities. We’ll be following up here with more details about the features above and posts about other improvements, so stay tuned!
Any news on metadata journaling? I had read previously it was intended to make it into version 1.10.
Unfortunately, journaling is not a feature in the 1.10.0 release, although it’s possible for a future release. Please stay tuned!
When will swmr be available on windows? As far as I know windows is not posix compliant.
Hi Stian – the SWMR feature itself works on Windows if SWMR writer and readers use local NTFS Windows file system. SWMR will not be supported on Windows in 1.10.0 since we do not have an automated testing on Windows. We will rework our documentation and will not use “POSIX compliant” requirement replacing it with the file system that has to provide consistent ordering of I/O operation as POSIX requires: “POSIX I/O writes must be performed in a sequentially consistent manner.”
Will you be adding options for relative path handling for external datasets in this release, so they can be resolved relative to the HDF5 file? This would make this feature much easier to use, particularly in cases where the external files are stored alongside the HDF5 file, and when the complete set of files is exchanged between users with different directory structures. For example, I want to record some data on one machine, and give it to other users elsewhere for analysis.