Our non-profit mission is to ensure efficient and equitable access to science and engineering data across platforms and environments, now and forever.
The HDF Group develops and maintains the feature-packed HDF5® technology suite, which is backed by complementary services. While help is available in the vast ecosystem, our experience and expertise will give you the decisive edge.
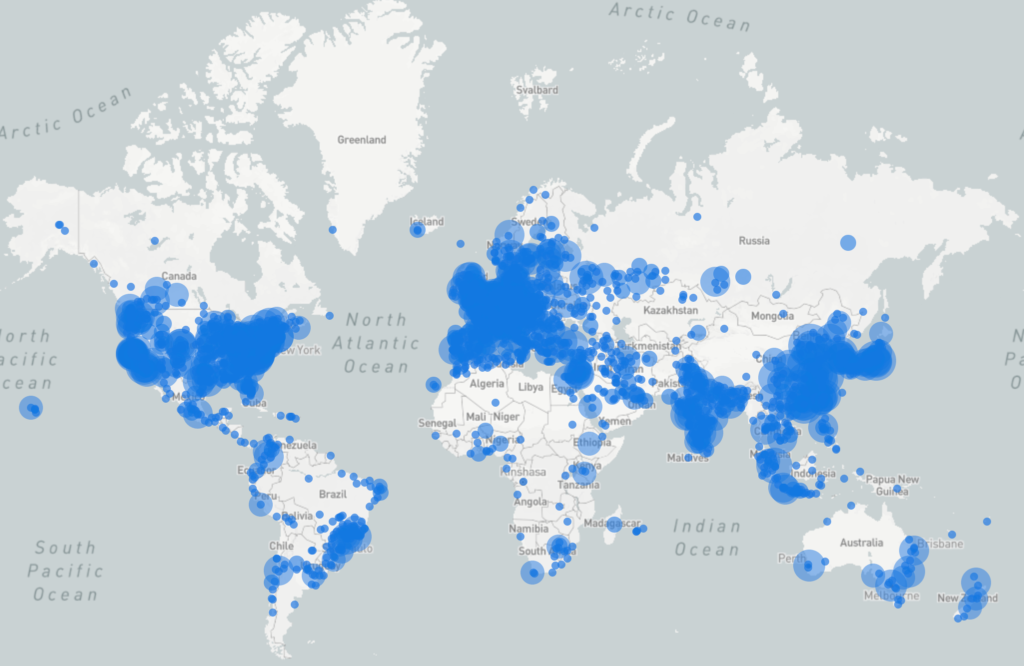
For over 25 years, people worldwide have relied on our open-source HDF software to solve some of the most challenging data management problems.
Weekly HDF Clinic
Learn more about Call the Doctor, our weekly session where our experts will present some information on their work and answer community questions.
