Rick, HDFql team, HDF guest blogger
HDFql (Hierarchical Data Format query language) was recently released to enable users to handle HDF5 files with a language as easy and powerful as SQL.
By providing a simpler, cleaner, and faster interface for HDF across C/C++/Java/Python/C#, HDFql aims to ease scientific computing, big data management, and real-time analytics. As the author of HDFql, Rick is collaborating with The HDF Group by integrating HDFql with tools such as HDF Compass, while continuously improving HDFql to feed user needs.
Introducing HDFql
If you’re handling HDF files on a regular basis, chances are you’ve had your (un)fair share of programming headaches. Sure, you might have gotten used to the hassle, but navigating the current APIs probably feels a tad like filing expense reports: rarely a complete pleasure!
used to the hassle, but navigating the current APIs probably feels a tad like filing expense reports: rarely a complete pleasure!
If you’re new to HDF, you might seek to avoid the format all together. Even trained users have been known to occasionally scout for alternatives. One doesn’t have to have a limited tolerance for unnecessary complexity to get queasy around these APIs – one simply needs a penchant for clean and simple data management.
This is what we heard from scientists and data veterans when asked about HDF. It’s what challenged our own synapses and inspired us to create HDFql. Because on the flip-side, we also heard something else:
- HDF has proven immensely valuable in research and science
- the data format pushes the boundaries on what is achievable with large and complex datasets
- and it provides an edge on speed and fast access which is critical in the big data / advanced analytics arena
With an aspiration of becoming the de facto language for HDF, we hope that HDFql will play a vital role in the future of HDF data management by:
- Enabling current users to arrive at (scientific) insights faster via cleaner data handling experiences
- Inspiring prospective users to adopt the powerful data format HDF by removing current roadblocks
- Perhaps even grabbing a few HDF challengers or dissenters along the way…
A simpler, cleaner and faster interface for HDF
We set out with the humble intention to offer the community a clean (yet powerful) way of managing HDF data. Our North Star was to aim for a solution similar to SQL. As a result, users can now handle HDF data through a high-level language that allows them to focus on solving real problems rather than spending valuable time dealing with low-level implementation details or endless lines of code.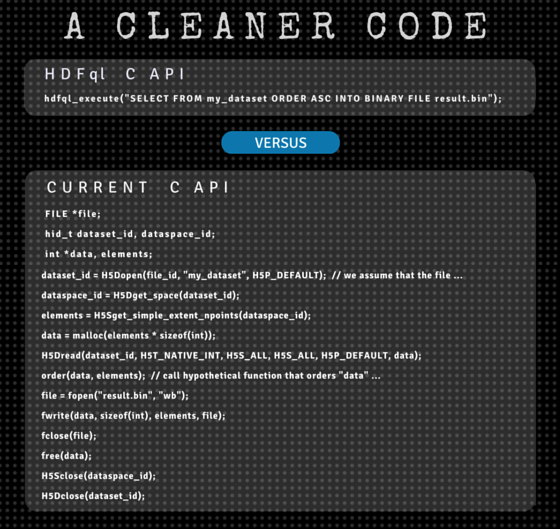
To exemplify how HDFql makes data management cleaner and simpler, let’s walk through a few important regions of its cortex (read “operations”):
- CREATE – create HDF files, groups, datasets, attributes, and (soft/hard/external) links.
- INSERT – insert (i.e. write) data into datasets and attributes. The input source can be direct values, cursors, user-defined variables, or (text/binary) files.
- SELECT – select (i.e. read) data from datasets and attributes. The output source can be cursors, user-defined variables, or (text/binary) files.
- SHOW – Introspect the content of HDF files such as get all existing datasets in a group, get the datatype of an attribute, etc.
To demonstrate how these cortical operations work in practice, nothing better than giving a scenario a spin and solving it through HDFql!
Imagine that we are acquiring data from a process which returns an array of 1024 integers on each reading. Additionally, let’s imagine that we do not know beforehand how many arrays will be returned by this process, and that we are required to store the data in HDF using the least amount of storage space possible. One solution would be to use a compressed extendible dataset and hyperslab features to store the acquired data. In C/C++ and using HDFql, this solution could easily be implemented as follows (note that we do not include comments in the code snippet since HDFql operations are basically self-explanatory.)
char script[100];
int data[1024];
int row;
row = 0;
hdfql_execute("CREATE FILE my_file.h5");
hdfql_execute("USE FILE my_file.h5");
hdfql_execute("CREATE CHUNKED DATASET my_dataset AS INT(UNLIMITED, 1024) ENABLE ZLIB");
hdfql_variable_register(&data);
while (acquire(data))
{
sprintf(script, "INSERT INTO my_dataset(%d) VALUES FROM MEMORY %d", row++, hdfql_variable_get_number(&data));
hdfql_execute(script);
hdfql_execute("ALTER DIMENSION my_dataset TO (+1)");
}
hdfql_variable_unregister(&data);
hdfql_execute("CLOSE FILE");Users familiar with other APIs for HDF should instantly realize how much clearer – and less verbose – it is to solve the above scenario with HDFql in C/C++ (for illustrations of other programming languages, see hdfql.com/examples).
Solving HDF challenges of today’s organizations
Nowadays, organizations usually work with more than one programming language. This demands resources and can be cumbersome for developers who must learn how to handle HDF data in each of these. Even if there are good solutions for individual programming languages, this does not solve the problem of operating HDF across disparate languages. With HDFql, this is part of the past since HDFql abstracts users from all intrinsic details and specificities of programming languages.
To illustrate this point, let’s go back to our previous scenario. Imagine that we are now required to read the entire dataset and write it in an ascending order into a binary file named “result.bin”. In C/C++ and using HDFql, this could be implemented through one simple statement:
// New HDFql C/C++ API
hdfql_execute("SELECT FROM my_dataset ORDER ASC INTO BINARY FILE result.bin");To implement the equivalent in Java, Python and C#, the statements are equally simple (and syntactically similar):
// New HDFql Java API
HDFql.execute("SELECT FROM my_dataset ORDER ASC INTO BINARY FILE result.bin");
// New HDFql Python API
HDFql.execute("SELECT FROM my_dataset ORDER ASC INTO BINARY FILE result.bin")
// New HDFql C# API
HDFql.Execute("SELECT FROM my_dataset ORDER ASC INTO BINARY FILE result.bin");As one can see, in HDFql not much changes between these programming languages except the case of the function we are calling (i.e. ’execute’). Great news for developers, especially when compared with alternative APIs:
// Current C API
FILE *file;
hid_t dataset_id, dataspace_id;
int *data, elements;
dataset_id = H5Dopen(file_id, "my_dataset", H5P_DEFAULT); // we assume that the file was opened previously
dataspace_id = H5Dget_space(dataset_id);
elements = H5Sget_simple_extent_npoints(dataspace_id);
data = malloc(elements * sizeof(int));
H5Dread(dataset_id, H5T_NATIVE_INT, H5S_ALL, H5S_ALL, H5P_DEFAULT, data);
order(data, elements); // call hypothetical function that orders "data" in an ascending way
file = fopen("result.bin", "wb");
fwrite(data, sizeof(int), elements, file);
fclose(file);
free(data);
H5Sclose(dataspace_id);
H5Dclose(dataset_id);
// Current Java API
DataOutputStream file = new DataOutputStream(new FileOutputStream("result.bin"));
int data[][], datasetID, dataspaceID, i, j, rows;
datasetID = H5.H5Dopen(fileID, "my_dataset", HDF5Constants.H5P_DEFAULT); // we assume that the file was opened previously
dataspaceID = H5.H5Dget_space(datasetID);
rows = H5.H5Sget_simple_extent_npoints(dataspaceID) / 1024;
data = new int[rows][1024];
H5.H5Dread(datasetID, HDF5Constants.H5T_NATIVE_INT, HDF5Constants.H5S_ALL, HDF5Constants.H5S_ALL, HDF5Constants.H5P_DEFAULT, data);
order(data); // call hypothetical function that orders "data" in an ascending way
for(i = 0; i < rows; i++)
for(j = 0; j < 1024; j++)
file.writeInt(data[i][j]);
file.close();
H5.H5Sclose(dataspaceID);
H5.H5Dclose(datasetID);
# Current Python API
dset = f["my_dataset"] # we assume that the file was opened previously
data = np.sort(dset) # call NumPy method to order content in an ascending way
data.tofile("result.bin") # call NumPy method to write "data" into binary file "result.bin"
// Current C# API
BinaryWriter file = new BinaryWriter(new FileStream("result.bin", FileMode.Create));
H5DataSetId datasetID;
H5DataSpaceId dataspaceID;
int [,]data;
int i, j, rows;
datasetID = H5D.open(fileID, "my_dataset"); // we assume that the file was opened previously
dataspaceID = H5D.getSpace(datasetID);
rows = H5S.getSimpleExtentNPoints(dataspaceID) / 1024;
data = new int[rows, 1024];
H5D.read(datasetID, H5T.H5Type.NATIVE_INT, new H5Array(data));
Order(data); // call hypothetical function that orders "data" in an ascending way
for(i = 0; i < rows; i++)
for(j = 0; j < 1024; j++)
file.Write((int) data[i, j]);
file.Close();
H5S.close(dataspaceID);
H5D.close(datasetID);Coding headache, anyone?
A side note (and added benefit): when executing HDFql operations, parallelism is automatically employed – whenever possible – to speed up operations. This means that all CPU cores available are used without developers having to worry about it (in the above HDFql snippet, this happens when ordering the dataset ascendingly).
Let’s walk through a few other features…
HDFql key (and future) features
HDFql currently offers the key benefits of:
- Offering a one stop high-level language across programming languages (C/C++, Java, Python and C#)
- Supporting several platforms (Windows, Linux and Mac OS)
- Reading and writing HDF data
- Supporting chunked/extendible datasets
- Supporting hyperslab functionalities
- Supporting variable-length datatypes
- Using automatic parallelism whenever possible
In upcoming releases, we plan to add even more power to HDFql by:
- Supporting H5Q and H5X APIs (querying and indexing features).
We are in touch with The HDF Group and as soon as these APIs are released for production, HDFql SELECT and SHOW operations will be extended.
- Supporting BLAS operations on the fly.
Operations such as this one will soon become a reality with HDFql:
SELECT FROM my_dataset_0 * my_dataset_1
Here, HDFql reads both datasets “my_dataset_0” and “my_dataset_1” and multiplies these using all available CPU/GPU cores (thanks to MAGMA, a BLAS API) before returning the result to the user.
- Supporting compound and enumeration datatypes.
Users will be able to easily create datatypes like this:
CREATE DATATYPE my_compound AS COMPOUND(my_field_0 AS DATATYPE_0, ..., my_field_X AS DATATYPE_X)
CREATE DATATYPE my_enumeration AS ENUMERATION(my_field_0 [AS my_value_0], ..., my_field_X [AS my_value_X])
- Supporting access to distributed (remote) data stored in HDF REST and Hyrax OPeNDAP servers.
- Supporting Fortran and R.
Making HDFql work for you
In the end, we are doing this for the appreciation of data and simplifying its management. On a personal note, at the HDFql team we are especially passionate about Climate and Earth Science. Knowing the historic role HDF has played in these fields, we truly hope that HDFql can contribute to faster future discoveries in these realms. Overall, if the broad community of HDF users finds reward in the simplified data management experience of HDFql, it makes our years of coding all worthwhile!
We hope with this intro to have given a flavor of how HDFql can aid you as a user of HDF. We are learning with the community along the way and strongly encourage user-driven insights to shape HDFql – so do not hesitate to reach out to us with key needs/priorities for your organization, suggested features, or other feedback.
Stay tuned for powerful releases of HDFql and news on upcoming integrations with key HDF Group tools. Here’s to managing HDF data simpler, cleaner, and faster!
HDFql is available at hdfql.com; HDFql is distributed as freeware. It may be used free of charge both for commercial and non-commercial purposes.
For news and updates, follow HDFql at twitter.com/hdfql
Rick can be reached directly at rick@hdfql.com
Editor’s Note: I’m really excited about Rick’s contribution to the HDF community — HDFql — which directly tackles many of the items on users’ wishlists for HDF5; most notably: 1. compact code, 2. easy learning curve (ideally something familiar), and 3. portability across platforms and languages. I encourage everyone — particularly those with SQL background — to check it out and provide feedback. Dave Pearah, The HDF Group, CEO
Please clarify the software license for HDFql? https://support.hdfgroup.org/products/hdf5_tools/SWSummarybyType.htm lists it as “open source”.
Hi Scott,
I reached out to Rick of HDFql regarding your question. He had me update the HDF5 Tools page, which is now maintained at https://portal.hdfgroup.org/display/support/Software+Using+HDF5 HDFql is now listed as freeware. Please let The HDF Group, or Rick of HDFql (email in the blog post) know if you have any more questions.
Thanks!