John Readey, The HDF Group
When using HDF5 or HSDS you’ve likely benefited (even if you weren’t aware of it) caching features built into the software that can drastically improve performance. In general, a cache is a software or hardware component that stores recently accessed data in a way that it is more efficient to access—computer memory versus disk, for example. The HDF5 library utilizes two types of caching: a chunk cache and a metadata cache that speeds up access to (respectively) dataset values and HDF5 metadata… Similarly, HSDS and h5pyd utilize caching to improve performance for service-based applications. In this post, we’ll do a quick review of how HDF5 library caching works and then dive into HSDS and h5pyd caching (with a brief discussion of web caching).
Caching in the HDF5 Library
To illustrate how the caching works in the HDF5 library, let’s consider a concrete example. Consider a HDF5 dataset with dimensions (5000 x 256) of type float32 that uses gzip compression. Since we are using compression, a chunk layout is required. The chunk layout determines how the library will store dataset data within a file and is set at dataset creation time.
In our example if the chunk layout is (1024, 256), the dataset values are stored in up to 5 distinct areas within the file (which may or not be contiguous based on how the data was written). By storing data in chunks data can be compressed/uncompressed on a chunk by chunk basis rather than having to read the entire dataset into memory.
Consider a Python code snippet:
for i in range(5000):
arr = dataset[i,:]
print(arr.mean())
In the first 1024 iterations of this loop, the same chunk is accessed by the dataset[i,:] selection (returning the ith row of the dataset). To return this selection, the library will need to read the entire chunk from disk, uncompress it and return the ith row of the chunk to the caller. It’s easy to see that if the library had to read the same data from disk and uncompress it for each loop iteration, it would slow things down quite a bit – though the computer’s disk caching (more caching!) would likely help with the disk reading part at least.
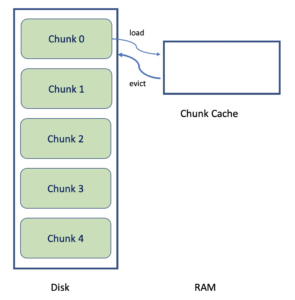
Enter the chunk cache! The chunk cache is a memory buffer that the library uses to store recently accessed chunks. In the code snippet, in the first iteration of the loop, chunk zero would be read from disk, uncompressed, and stored in the chunk cache. The library will then return the indicated selection to the caller.
The payoff comes in the second loop iteration… The library will see that chunk zero is resident in the cache and can directly return the selection [1,:] rather than going back to disk. This happy state of affairs will continue until iteration 1024 in which case chunk one is read from disk, stored in the cache and selection [1024,:] is returned. Then we’ll see another series of cache hits for iterations 1024 through 2047 and so on.
Where does the metadata cache fit in? This cache is used to store recently accessed metadata (e.g. dataset properties like shape, type, compression filter settings). This metadata is stored in the file separately from the dataset data but the metadata will be needed by the library to perform any dataset selection. By keeping recently accessed metadata in memory we avoid yet another disk i/o operation.
Going back to the chunk cache, how can things go wrong? One thing to be aware of is that the chunk cache is allocated per dataset and is a fixed size—1 MiB by default. That works fine for our example since we only access one chunk at a time and the chunk’s size is 1MiB (1024 x 256 x 4). What if we create a dataset with the same shape, and chunk layout but use float64 as the type? Now we have 2 MiB chunks which won’t fit in the chunk cache. So in this case each iteration of the loop will require a disk I/O operation followed by uncompression, slowing things done a bunch.
Fortunately you can choose a chunk cache size when you open a dataset. Assuming memory is not severely constrained, you’d want to specify a chunk cache size of at least 2 MiB. In h5py you can override the default chunk cache size when opening a file by using the “rdcc_nbytes” parameter and giving the desired size in bytes. See the documentation for the specifics.
Even when the chunk cache is large enough to hold a given chunk, what happens when multiple chunks are accessed? If a new chunk is accessed, but the chunk cache is full, one or more of the existing chunks will need to be evicted—removed from the cache to make room for the incoming chunk. This is no problem in our example since we only need access to one chunk at a time and once we’ve moved beyond a given chunk we never return to it.
But consider we tweak our example so that we are accessing data by columns rather than rows:
for i in range(256):
arr = dataset[:,i]
print(arr.mean())
Now in each iteration we are accessing all 5 chunks. This will lead to a constant series of chunk evictions and negate the value of having the cache. (You could argue that a chunk shape like (5000 x 1) would be better suited for this application, but you don’t always have the freedom to define the chunk layout—e.g. you are reading a file created by another application).
In this case we’d want a chunk cache size of at least 5 MiB (still quite reasonable for most systems). In our example this will result in all the dataset data being read into memory on the first iteration and then being available for each future iteration.
This has been a high-level overview of how chunk and metadata caching work in the HDF5 library. See Chunking in HDF5 for a more in-depth description.
Caching in HSDS
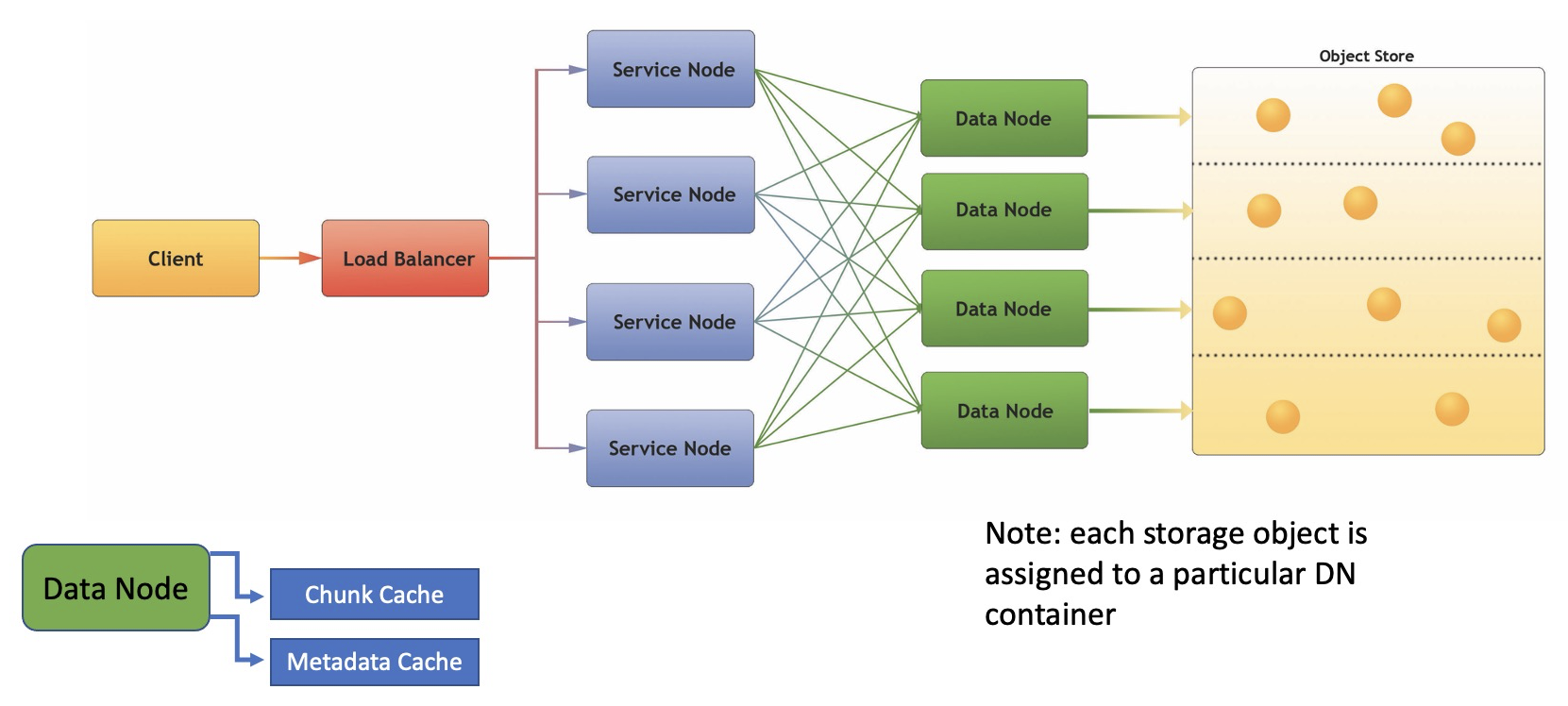
Similar to the HDF5 library, HSDS maintains both a metadata and a chunk cache. But while in the HDF5 library the caches are created when a file is opened and are destroyed when the file is closed, in HSDS the caches persist for as long as the service is running.
Consider the following python script:
import h5pyd as h5py
f = h5py.File("/nrel/wtk-us.h5", bucket="nrel-pds-hsds")
dset = f["windspeed_80"]
arr = dset[522, ::, ::]
print(arr.mean())
Here we retrieve slice 522 from a 3-dimensional dataset (about 18 MiB of data) and print out the mean of the values returned. Unless the server is quite busy, the chunks used in the selection will be resident in the chunk cache if the program is run multiple times.
Running this program on HDF Lab, a quick experiment resulted in the first run taking 2.46 seconds while the second taking only 444 ms, so there’s quite a large benefit from being able to retrieve the data from the cache versus reading from S3 (AWS S3 latency is quite a bit higher than local disk latency, so the advantage of caching the data is relatively higher).
Since data is being cached on the server rather than the client, other clients (perhaps on other machines) will also benefit from recently accessed data being in the metadata or chunk cache, i.e. The cache is a shared resource for all the clients accessing that server.
Like the HDF5 library, metadata and chunk caches, the HSDS cache operates on a “least recently used” (LRU) principle—in the event the cache is full, the least recently accessed object is evicted from the cache.
But unlike the HDF5 library, the HSDS caches cannot be configured at file open time, but rather the size of the cache is determined by server configuration parameters and the number of nodes currently running in the service. For the metadata cache, the configuration parameter is metadata_mem_cache_size which defaults to 128 MiB. For the chunk cache the configuration parameter is chunk_mem_cache_size which also defaults to 128 MiB. These are the “per node” sizes. If you are running HSDS with 8 DN (data node) containers, then total cache size would be 128 MiB x 8 = 1 GiB.
These configuration parameters are quite conservative, and based on the amount of memory available, and the type of applications used with the server, it may be beneficial to use a larger value for these (the chunk_mem_cache_size in particular). Just be sure to increase the Docker or Kubernetes mem limits correspondingly so that the container will have enough memory to host the cache as well as its other memory needs.
Here it might be helpful to point out that HSDS is architected in such a way that any given object is “assigned” to a particular DN container based on its id (the assignment is computed by taking a hash of the object uuid). So it’s never the case that the same chunk would be stored in two DN containers. This property can be utilized if it happens that there is a certain set of data that is frequently accessed and it’s imperative that the latency to read the data be as low as possible. For example, consider a dataset of 100 GiB and you’d like to configure HSDS so it would be possible to have the entire dataset in memory at one time. In this case you could run 50 DN containers configured with a 2 GiB chunk cache. If the server used to host HSDS on Docker does not have sufficient memory to run that many containers, you could use Kubernetes to run HSDS across a cluster of machines. In any case, as clients accessed different chunks of the dataset over time it would be more likely that the chunk would be found in memory and the number of requests to the storage service in the long run would be minimal.
What happens if the number of DN containers changes as HSDS is running? With Docker the number of containers is typically fixed when the server is started, but with Kubernetes the number of containers (pods) can be scaled dynamically. Since the assignment of an object to a DN container is based on the number of DN containers, when HSDS is scaled, the entire chunk and metadata cache are invalidated. As objects are loaded from storage using the new assignment function that cache will get re-populated. So frequently scaling HSDS may not be desirable at least if you like to keep objects around in the cache as long as possible!
There are two other cache related configuration parameters that might be of interest: metadata_mem_cache_expire and chunk_mem_cache_expire both of which default to 3600 seconds (one hour). This configuration controls the maximum time an item can live in the cache even if it’s not evicted for other reasons. Why not have an object live in the cache for as long as the server is running? Consider the scenario where we have two HSDS instances running. One is used mainly to modify data and one which is used by clients to read data. They are both setup to read/write to the same bucket. The effect of the metadata and chunk cache is that clients of the second HSDS service may miss updates made by the first service. If a particular object is present in the cache, the modified data that has been written to storage won’t be visible.
The effect of the expire config, is that a cache item will always be invalidated if it’s been in the cache longer than the expire interval. Thus in our scenario, the read clients will see data that is delayed no longer than the expire interval. If a more real-time view is needed, this configuration can be lowered significantly with only a moderate decrease in performance. For very dynamic data, the simplest thing to do would be to have both the reader and writer clients use the same server (in which case the expire value wouldn’t matter), but for various reasons it can be convenient to set up multiple HSDS instances using the same storage bucket.
There’s yet another cache used in HSDS: the data cache. This is only used when HSDS is reading regular HDF5 files rather than data in the HSDS sharded format. We’ll explore how this works in a future blog post.
Caching in h5pyd
On the client-side, there’s no equivalent of the chunk cache with the Python HSDS client h5pyd. Typically it wouldn’t be efficient to read entire chunks from the server if the client just needs to access a small section of each chunk, so a chunk cache hasn’t been implemented.
There is a metadata cache though and this can be quite useful given that the latency between the client and the server can be quite high compared with assessing data on local disk. Consider a script that prints out each attribute of each object for a file (HSDS domain):
import h5pyd as h5py
def my_visit(name, obj):
print(f“attributes for {name}:”)
for k, v in obj.attrs:
print(f” {k}: {v}”)
f = h5py.File(file_path)
f.visititems(my_visit)
The function my_visit will be called once for each object, and each attribute of each object needs to be read. If each link iteration and each attribute iteration required a server roundtrip, that could be quite slow.
Instead, in response to http GET domain request (the REST call that h5pyd makes in the file open function), all metadata for the domain is returned as a single JSON response (unless that would be overly large—more than a few MB). The h5pyd package keeps this JSON in memory for as long as the reference to the domain is open. Therefore, in the script above you’ll typically see only the h5py.File(file_path) call taking significant time. Iterating through the domain objects or object attributes will take hardly any time since the needed data is already present on the client.
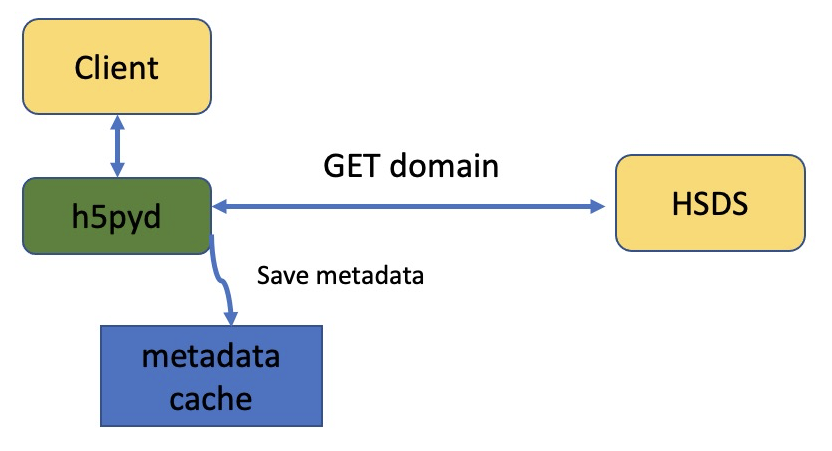
What if you are creating a script that will be used to read from a domain that is being frequently modified? (say, another process is periodically adding new attributes or links) In that case, you can set the use_cache parameter of File open to False:
f = h5py.File(file_path, use_cache=False)
This will cause h5pyd to not request the metadata in the GET request, but ask for link or attribute data as needed. So, if for example a link is added by another process, this will be seen by the reader process as soon as it lists the links in the group. This parameter is also useful to speed things up if you expect to open a file with a large amount of metadata, but don’t anticipate accessing much of it (e.g. you just need to read one attribute from the root group).
Web Caching
There’s one other type of caching that is not strictly part of HSDS or h5pyd (and which I’ve never actually used myself), but which might be useful for some use cases, which is web caching. It’s very common in web architectures to set up a proxy server between the client and web server. The client sends the same GET, PUT, POST, DELETE requests to the proxy as it would to the web server. If the proxy has seen a particular GET request before and has the response to the request in its cache, it returns the cached response. If not, it forwards the request to the web server, saving the response and returning the response to the client.
This is used frequently to lower the latency to servers that are far away from the client. By having a proxy that runs closer to the client (say in the same datacenter), latency can be reduced compared with each request having to go to the more distant web server.
Another use case is when the server is in danger of being overloaded. If a sizable fraction of requests can be handled by the proxy, that reduces the amount of work the server has to perform.
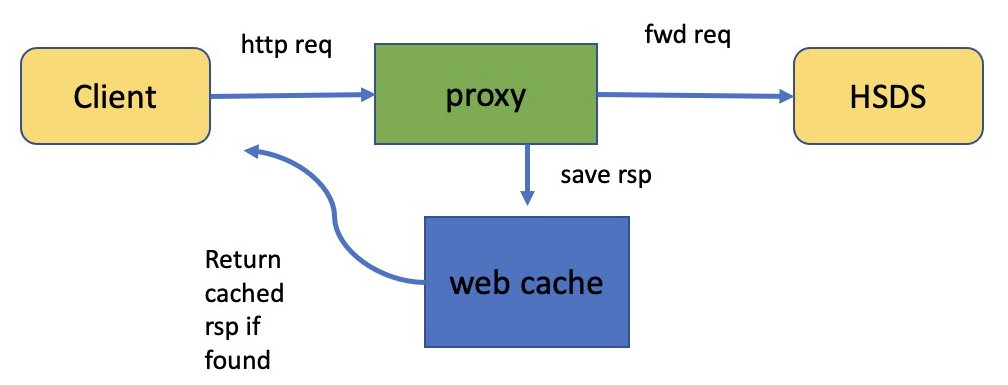
It would certainly be feasible to use a proxy server with HSDS—at least for scenarios where the data is relatively static. In fact it was the desire to support proxy servers that led to the HDF REST API being designed as it was. Consider the REST request to read the values from a given dataset in the domain /shared/tall.h5:
The dataset is referenced by its UUID rather than the h5path to the dataset (which would be “/g1/g1.1/dset1.1.1” in this case). Using the h5path might seem a more natural way to structure the REST API, but using the UUID has certain advantages. Consider that it’s quite possible for the same dataset to be referenced by multiple paths in HDF5. By using the object UUID in the REST request we have a canonical way to represent that HTTP resource. If multiple URLs are used to refer to the same resource, that would cause some difficulties for the proxy, as it would have no way to know that the two URLs actually refer to the same object.
If anyone has used a web cache with HSDS, we’d certainly be interested in hearing how it worked! Feel free to post any questions about caching to the HDF5 or HSDS forums (or as a comment to this post), and we’ll do our best to answer them.