by John Readey, The HDF Group
Accessing large data stores over the internet can be rather slow, but often you can speed things up using multiprocessing—i.e. running multiple processes that divvy up the work needed. Even if you run more processes than you have cores on your computer, since much of the time each process will be waiting on data, in many cases you’ll find things speed up nicely.
As an example, let’s look at NREL’s WTK CONUS dataset (see: https://www.nrel.gov/grid/wind-toolkit.html). This is a set of 8 HDF5 files stored on AWS S3 (in the us-west-2 region) providing data for the years 2007 to 2014. The files contain datasets for temperature, windspeed, wind direction, pressure, etc., stored as 2D datasets (dimensions time and location index). Each file is slightly more than 1 TB in size, so downloading the entire collection would take around a month with a 80 Mbit/s connection. Instead, let’s suppose we need to retrieve the data for just one location index, but for the entire time range 2007-2014. How long will this take?
| Wind Integration National Dataset Toolkit | Grid Modernization – NREL
Gridded Atmospheric WIND Toolkit. This 50-TB subset contains instantaneous 1-hour resolution model output data for 7 years on a uniform 2-km grid that covers the continental U.S., the Baja Peninsula, and parts of the Pacific and Atlantic oceans. www.nrel.gov |
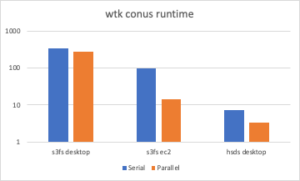
If we are using python, a simple script can open each file in turn, read from the dataset (either with h5py and s3fs or with h5pyd and the HSDS service), and fetch the desired data. You can find an example script here: https://github.com/HDFGroup/hsds/blob/master/tests/perf/nrel/wtk/wtk_conus.py. Running the command: python wtk_conus.py --folder=s3://nrel-pds-wtk/conus/v1.0.0/, on my Mac, this takes quite a while to run: 347 seconds.
Since most of this time is spend waiting on data requested from S3, we can speed things up by running multiple processes, one process for each year. In Python, the multiprocess module makes this easy. In the script we initialize the Pool class (https://docs.python.org/3/library/multiprocessing.html#multiprocessing.pool.Pool) with the number of processes desired. Then pool.map()will execute a given function once for each process.
 The
The --mp flag will tell our script to use multiprocessing, and with this (python wtk_conus.py --folder=s3://nrel-pds-wtk/conus/v1.0.0/ --mp) we get a better result: 278 seconds. Not an 8x speedup though since we’re limited in the bandwidth we can send over the public internet. If I run the script on an EC2 instance instead we get results of 99 seconds in serial mode and 14 seconds in parallel, close to linear speedup.
https://developer.nrel.gov/api/hsds, we can run: python wtk_conus.py --folder=hdf5://nrel/wtk/conus/ and the runtime was just 7.3 seconds. If we use the --mp option on top of that, the runtime is just 3.3 seconds.
So, we can see (at least in this example), some real benefits of using multiprocessing (and HSDS). For your own workloads, your milage will certainly vary, but I’d encourage everyone to explore multiprocessing and see how it might help.
A quick mention that there are two other means of concurrency that are commonly used in Python: multithreading and asyncio. Multithreading uses lighter weight “threads” rather than processes (but the programming model is very similar). Unfortunately threading doesn’t work well with h5py or h5pyd, in each case the thread will end up waiting on I/O of the other threads to complete, negating the benefit of using threads in the first place.
Asyncio is an approach using what is known as “Task-based parallelism”. In the asyncio model, tasks run until they are blocked (say on an I/O request) and give up control to another task. This approach is how HSDS implements parallelism internally. Currently though, neither h5py or h5pyd support async interfaces (though you can use asyncio with the HDF REST API).
Let us know if this or other techniques are useful for your projects!