Jordan Henderson
The HDF Group just released HDF5 1.13.1. All of the 1.13 series are experimental releases, which allows us to test new features with our users and get feedback while we are working on the development of the next major maintenance release.
Performance improvements and improved support for collective I/O
The main goal of working on HDF5’s parallel compression feature was to improve usability by:
- Fixing bugs that were impacting the stability of the feature
- Reducing memory usage of the feature
However, several performance enhancements were also made, including improved support for collective MPI I/O, reduction of overhead from MPI usage, better use of MPI non-blocking operations, several internal algorithm optimizations, and more. To get a sense of the impact of these changes, support for HDF5 dataset filters was added to IOR (not yet integrated with the main repository) and some initial results for the feature were gathered on Cori at Nersc:
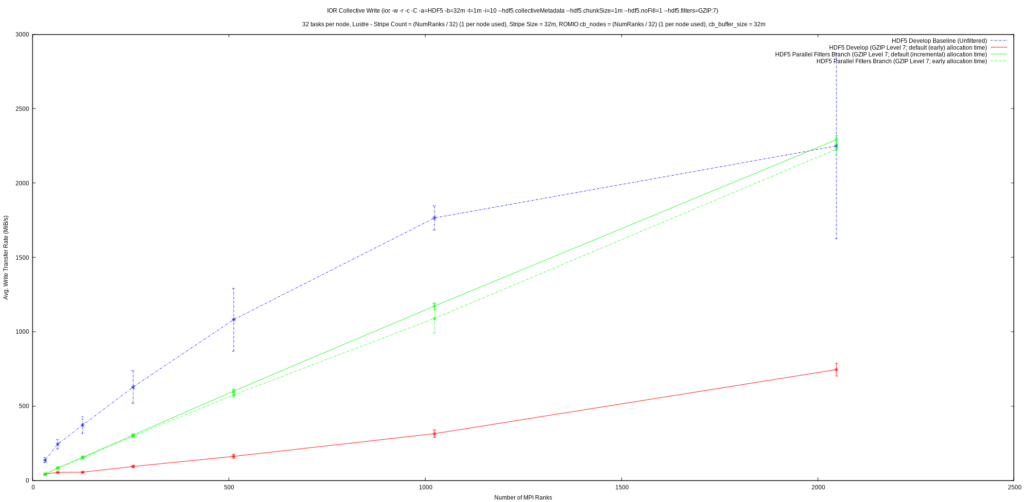
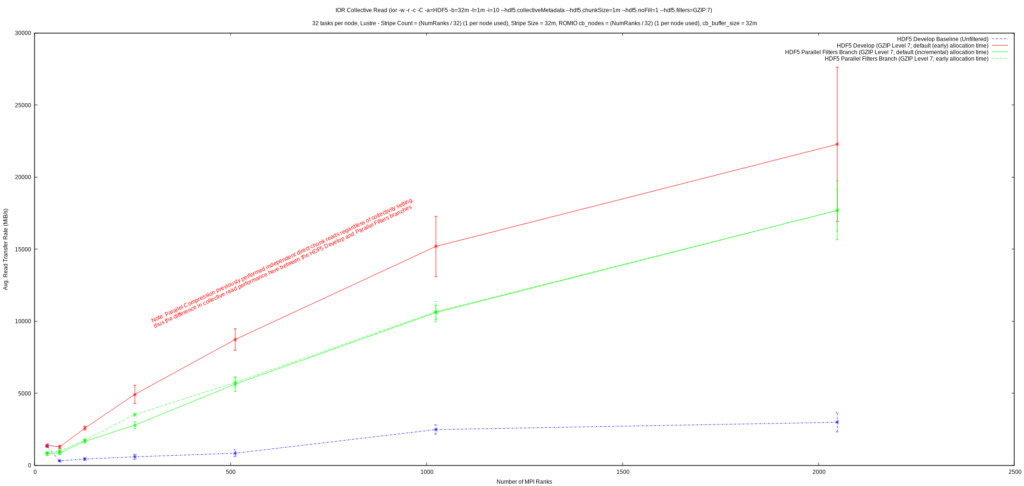
From the first graph, it can be seen that write performance was improved upon significantly and exhibits much better scaling than previously. A quick glance at the second graph reveals that collective read performance is a fair bit worse than previously, but it’s important to realize that the initial implementation of parallel compression in HDF5 always performed reads of dataset chunks independently, regardless of the collectivity setting from the application. This had the effect that:
- Collective reads of filtered datasets were always done independently
- Collective writes of filtered datasets would issue independent reads for chunks that needed reading before being modified and written back
The feature now correctly supports collective reads, but, as these graphs show, collective reads may not perform the best and HDF5 applications that perform reads of filtered datasets collectively may now experience worse read performance than before. The IOR results show that there may be some potential performance problems with collective dataset reads in HDF5 in general, whereas independent reads appear
to perform well. Correct support for collective I/O, along with these potential performance problems, likely explains this discrepancy in parallel compression’s collective read performance.
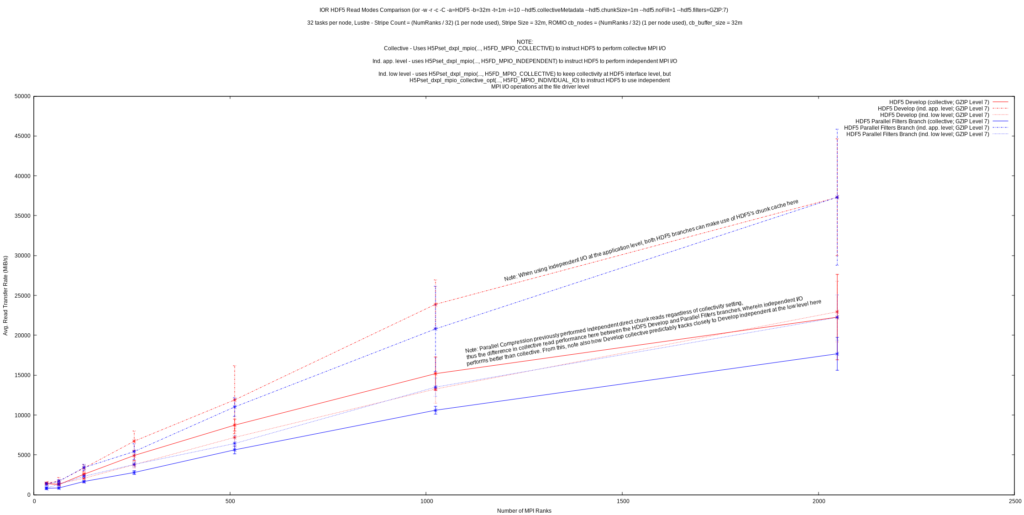
The final graph shown appears to confirm this by highlighting that the performance of collective and low-level independent reads in the HDF5 develop branch (before changes to parallel compression) track close to each other, as well as tracking close to low-level independent reads after the changes to parallel compression. For the time being, it is recommended for parallel HDF5 applications to perform dataset reads independently (parallel writes of filtered datasets are still required to be collective) until performance issues with collective reads in HDF5 can be further investigated.
Incremental file space allocation support
HDF5 file space allocation time is a dataset creation property that can have significant impacts on application performance, especially if the application uses parallel HDF5. When left unspecified by the application, serial HDF5 defaults to a space allocation time that generally makes sense based on the storage type (compact, contiguous, chunked, etc.) chosen for a dataset’s raw data. However, parallel HDF5 forces early file space allocation time (H5D_ALLOC_TIME_EARLY) for datasets created in parallel; any setting from the application is effectively ignored. This means that all space in the file for datasets created in parallel will get allocated during dataset creation. Since allocation of space in the file needs to be done collectively, forcing early allocation time facilitates future independent access to a dataset’s raw data by ensuring that all of the dataset’s file space is fully allocated; e.g., MPI ranks won’t need further coordination beyond dataset creation in order to write to the dataset. As an example, if a dataset’s space allocation time was set to ‘incremental’ (only allocate file space as dataset chunks are initially written to), independent access to the dataset’s raw data becomes trickier. The space for currently-unallocated dataset chunks would need to be allocated collectively before those chunks could be written to independently, thus requiring some collective coordination for an operation that was intended to be independent.
While early file space allocation simplifies matters here, it can also have some potentially non-obvious drawbacks. For one, allocating all file space for a dataset at creation time will generally mean that the larger your dataset is, the more noticeable the overhead during dataset creation will become. Further, adding HDF5 fill values into the mix means you may be writing out fill data across a large portion of a dataset that you were planning to overwrite anyway! Initial usage of the parallel compression feature clearly exhibited an overhead issue when creating filtered datasets with fill values in parallel. A typical workaround to speed up dataset creation was to set HDF5’s fill time to ‘never’ to skip writing of fill values, but this isn’t ideal if your application actually wants to use fill values.
With the latest changes to HDF5’s parallel compression feature, incremental file space allocation time is now additionally supported for (and the default for) datasets created in parallel only if they have filters applied to them. Early space allocation time is still the default (and only applicable allocation time) for datasets created in parallel that do not have filters applied to them and is still supported for filtered datasets as well. This new feature can be supported since parallel writes to filtered datasets (where file space allocation will occur) must be collective, so we are not in danger of MPI ranks missing out on a collective space allocation operation. This change should significantly reduce the overhead of creating filtered datasets in parallel HDF5 applications and should help with applications that wish to use fill values on filtered datasets, but would typically avoid doing so since dataset creation in parallel would often take an excessive amount of time. Now, with incremental file space allocation support, allocation of file space for dataset chunks and writing of fill values to those chunks will be delayed until each individual chunk is initially written to, unless the application specifically requests early file space allocation time for a dataset.
Additional improvements in HDF5 1.13.1
Further improvements to HDF5’s parallel compression feature include the following:
- Significant reduction of memory usage for the feature as a whole
- Reduction of copying of application data buffers that are passed to
H5Dwrite - Addition of proper support for HDF5 dataset fill values with parallel compression
- Addition of support for HDF5’s flag to signal that partial chunks shouldn’t have filters applied to them
- Addition of an
H5_HAVE_PARALLEL_FILTERED_WRITESmacro toH5pubconf.hso HDF5 applications can detect at compile-time if the feature is enabled - Addition of simple examples (ph5_filtered_writes.c and
ph5_filtered_writes_no_sel.c) to demonstrate basic usage of the feature
As a reminder, our next maintenance release, HDF5 1.14.0 is targeted for release in late 2022 and will integrate the improvements made through our work on the experimental release 1.13. To make HDF5 1.14.0 the best for our users, we would appreciate your feedback (both positive and negative) on the HDF5 1.13.x releases and how they perform for your application. Please send your feedback to help@hdfgroup.org, or feel free to start a discussion on the forum.
This work was supported by the U.S. Department of Energy under Contract Number DE-AC52-07NA27344.